Imaging Collection
Imaging datasets play a crucial role in the research of neurological disorders, offering invaluable insights into the structure and function of the brain.
Imaging techniques like magnetic resonance imaging (MRI), positron emission tomography (PET), and functional MRI (fMRI) provide non-invasive means to visualize these changes, allowing researchers to investigate the underlying mechanisms and develop effective diagnostic and therapeutic strategies.
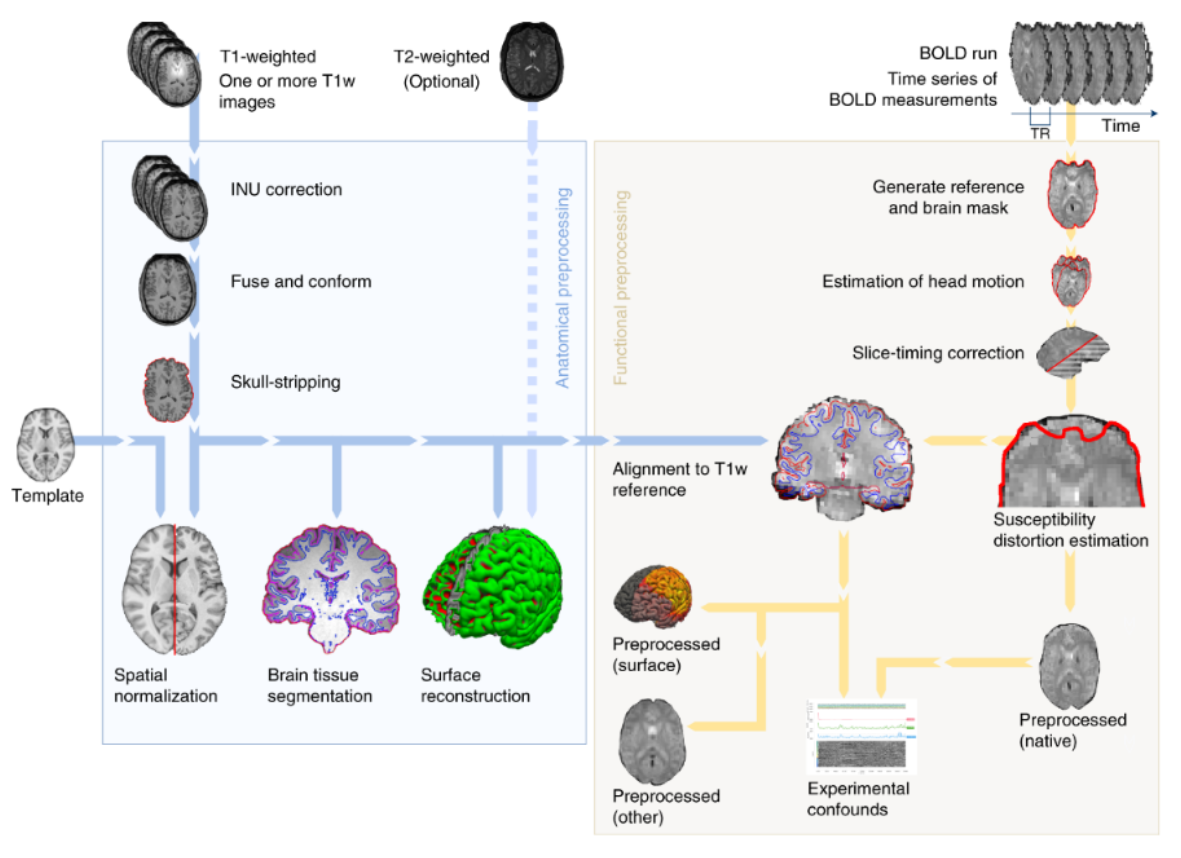
The DPUK Data Portal brings together raw imaging and image derived phenotypic data from over 3000 participants across 20 cohort studies. The raw imaging data is ingested into our imaging processing pipeline that defaces the images and standardises them to BIDs format to create research ready datasets.
For Data Owners
- A secure environment where your data never leaves
- Tools for anonymisation to deface scanse and remove Personal Identifiable Information
- Tagging of files to allow access to filtered data to researchers
- Processing pipelines to create research-ready datasets
For Researchers
- An analysis environment with access to CPU/GPU resources via HPC
- Windows and Linux environments available with different sizes according to need
- Efficient storage solutions for processing large volumes of data
- Access to a range of neuroimaging tools and software
Sharing Data
If you have collected a neuroimaging dataset, whether that's MRI, PET, MEG or anything else, and want to share it for other researchers to use, then you can share it on our secure research platform where you maintain in control of your data. Your data can never leave our environment and we have built robust ingest pipelines for anonymisation and processing to make sure your data is research-ready.
So how does our ingest pipeline work?
Submission
You can upload your data into our platform via a simple Secure File Transfer Protocol (SFTP) tranfer.
Data Check
Your data goes through our data check process to assess the state of the data to determine what level of anonymising and processing needs to happen.
Anonymisation
If your data contains PII either within any phenotypic files or image files then we will remove these to avoid any potential disclosure risk. We will also deface all structural scans.
Preprocessing
The data will then go through our processing pipelines to generate Imaging Derived Phenotypes (IDPs) along with processed scans so researchers have a choice of what they want to access.
Tagging
Data will then be tagged and stored in a bucket in our object storage server to allow for easy filtering of data to researchers and allows you to put controls on your data.
Analysis Environment
There are a choice of Windows and Linux desktops with varying sizes of memory and CPU count depending on your project requirements. Both of these environments have tools preinstalled to allow you to carry out all of the processing and analysis you need with the tools you are most comfortable with.
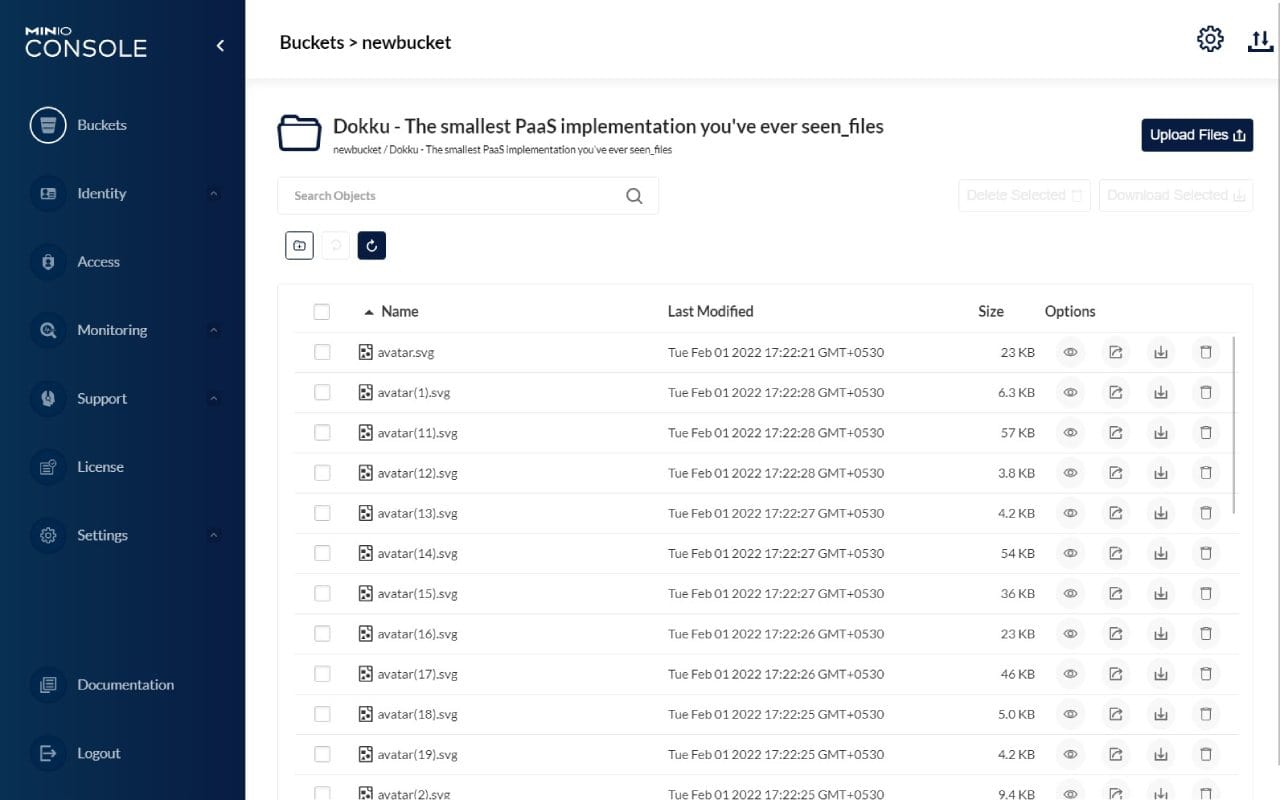
Bucket Storage
Our MinIO Object storage server allows data to be stored in buckets which can be accessed directly through the Python API. This allows us to store big data efficiently which is why we allow you up to 2.5 times the amount of storage for the data you are analysing.
Alternatively, data can be mounted to a drive if you want to access data in the typical file store system.
Processing Pipelines
All of our datasets come standardised to the BIDS format and contain derivates generated from our preprocessing pipeline. Whether you are a researcher or a data owner, we take away the lengthy process of generated derived data, and produce datasets which are research-ready.